Volume 19, Issue 1 (Jan-Feb 2025)
mljgoums 2025, 19(1): 6-8 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Livani F, Koohsar F, Tohidi F, Sharbatkhori M, Faridnia R, Yadagiri G, et al . Molecular evaluation of Leishmania species in negative ulcer smears from patients suspected of having cutaneous leishmaniasis referred to the health Center of Aq Qala city during 2019-2020. mljgoums 2025; 19 (1) :6-8
URL: http://mlj.goums.ac.ir/article-1-1715-en.html
URL: http://mlj.goums.ac.ir/article-1-1715-en.html
Fatemeh Livani1 

 , Faramarz Koohsar2
, Faramarz Koohsar2 

 , Farideh Tohidi2
, Farideh Tohidi2 

 , Mitra Sharbatkhori2
, Mitra Sharbatkhori2 

 , Roghiyeh Faridnia2
, Roghiyeh Faridnia2 

 , Ganesh Yadagiri3
, Ganesh Yadagiri3 
 , Ayeneh Hajieh Pangh2
, Ayeneh Hajieh Pangh2 

 , Mehdi Khoshrou1
, Mehdi Khoshrou1 

 , Hamed Kalani4
, Hamed Kalani4 




 , Faramarz Koohsar2
, Faramarz Koohsar2 

 , Farideh Tohidi2
, Farideh Tohidi2 

 , Mitra Sharbatkhori2
, Mitra Sharbatkhori2 

 , Roghiyeh Faridnia2
, Roghiyeh Faridnia2 

 , Ganesh Yadagiri3
, Ganesh Yadagiri3 
 , Ayeneh Hajieh Pangh2
, Ayeneh Hajieh Pangh2 

 , Mehdi Khoshrou1
, Mehdi Khoshrou1 

 , Hamed Kalani4
, Hamed Kalani4 


1- Infectious Diseases Research Center, Golestan University of Medical Sciences, Gorgan, Iran
2- Laboratory Sciences Research Center, Golestan University of Medical Sciences, Gorgan, Iran
3- Department of Veterinary Biosciences, The Ohio State University, Columbus, OH, USA
4- Infectious Diseases Research Center, Golestan University of Medical Sciences, Gorgan, Iran ,hamed.kalani@yahoo.com
2- Laboratory Sciences Research Center, Golestan University of Medical Sciences, Gorgan, Iran
3- Department of Veterinary Biosciences, The Ohio State University, Columbus, OH, USA
4- Infectious Diseases Research Center, Golestan University of Medical Sciences, Gorgan, Iran ,
Keywords: Leishmaniasis, Cutaneous, Leishmania major, Leishmania tropica, Negative Ulcer Smear, Multiplex Nested PCR
Full-Text [PDF 369 kb]
(239 Downloads)
| Abstract (HTML) (933 Views)
Conclusion
The results of this study showed the superiority of the multiplex nested PCR method over the microscopic method. In cases suspected of having CL, multiplex nested PCR can be used to avoid inappropriate treatment for patients where the samples are reported negative microscopically. Using the multiplex nested PCR method to determine the type of parasite that causes CL can provide researchers with more accurate information about the prevalence of the disease in epidemiological studies, which helps in making preventive decisions.
Acknowledgement
None.
Funding sources
This research was financially supported by the Vice Chancellor of Research at Golestan University of Medical Sciences. (Grant No: 112237).
Ethical statement
The Ethics Committee of Golestan University of Medical Sciences (Ethical code: IR.GOUMS.REC.1400.166) approved this study. This article has not been published in other journals, nor will it be, and is not under review in any journal.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Author contributions
Fatemeh Livani: Study design; Faramarz Koohsar and Mehdi Khoshrou: Sample collection; Farideh Tohidi: Original draft preparation; Mitra Sharbatkhori, Roghiyeh Faridnia, Ayeneh Hajieh Pangh: Laboratory work; Hamed Kalani: Supervision, Study design, Reviewing and editing; Ganesh Yadagiri: Critical revision.
Data availability statement
All data analyzed in this study are presented in this published article.
Full-Text: (19 Views)
Introduction
Cutaneous leishmaniasis (CL) is a prevalent infectious zoonotic disease between humans and animals, which is caused by different species of the genus Leishmania. This disease is transmitted to a new host by an infected mosquito bite (1). According to the reports of the General Department of Care and Prevention of Diseases, every year, about 20,000 cases of CL are reported in Iran. It is estimated that the actual number of cases is 6-10 times more than that which is reported (2). Golestan Province is one of the important centers of CL in Iran, which is mostly reported from rural populations and the margin of cities (3).
The prevention and control of CL varies depending on the species of the parasite. In endemic areas where the prevalence of CL is high, the correct and timely diagnosis of this disease is of particular importance. Thus, the use of diagnostic methods with high sensitivity and specificity such as PCR is suggested (4). PCR technique is considered a valuable method in epidemiological studies (5). In comparison, current methods for detecting CL such as microscopic examination of Giemsa-stained smears may report as false negative, especially in low parasitic burden (6). Misdiagnosed cases may lead to subsequent consequences for the patient, such as a delay in healing the lesions and the spread of the disease in the region (7).
Therefore, this study aimed to examine Leishmania species in negative ulcer smears from patients suspected of having CL referred to Aq Qala Health Center, Golestan Province, northeastern Iran, using the multiplex nested PCR method.
Methods
This retrospective cross-sectional study was performed on 72 negative ulcer smears from patients suspected of having CL referred to the health center of Aq Qala City from August 2019 to April 2020. These smears had been prepared by Giemsa stain and examined under a light microscope at 400 and 1,000 magnifications. The examiner's final report was negative for detecting Leishmania species.
DNA was extracted from the smears using a Blood DNA isolation kit (DENA Zist Asia, Mashhad, Iran) according to the manufacturer's instructions. The quantity and quality of the extracted DNA were analyzed using a NanoDrop ND-1000 spectrophotometer (PeqLab, Erlangen, Germany).
PrimerPlex v2.6 software was used to design primers for multiplex nested PCR. Two pairs of primers were used for each studied gene. Accordingly, the product of the first reaction was considered as the target of the second reaction. Therefore, two pairs of primers, one for the first reaction and the other for the second reaction, were considered for each of the examined parasites, Leishmania major and Leishmania tropica. Primers were designed for the SSU rDNA gene of the mentioned parasites. Furthermore, DNA for fragments 3 and 4 of histone protein was used to evaluate the accuracy of the multiplex nested PCR reaction as an internal control. The specifications of the primers used are given in Tables 1 and 2.
Multiplex TEMPase 2x Master Mix (Ampliqon, Denmark) was used for this aim according to the manufacturer's instructions. The reactions were performed in a 50 µL final volume, including 25 µL of the 2x Master Mix, 1 µL of each primer, target DNA, and PCR-grade H2O to a total reaction volume of 50 µL. Reaction 1 included 3 steps as follows: 1) denaturation at 94 °C for 2 min, 2) 35 cycles including denaturation at 94 °C for 15 s, annealing at 52.2 °C for 30 s, extension at 72 °C for 30 s, 3) final extension at 72 °C for 2 min. Reaction 2 was carried out in the same way as Reaction 1, except that the annealing was carried out at 50.3 °C.
Cutaneous leishmaniasis (CL) is a prevalent infectious zoonotic disease between humans and animals, which is caused by different species of the genus Leishmania. This disease is transmitted to a new host by an infected mosquito bite (1). According to the reports of the General Department of Care and Prevention of Diseases, every year, about 20,000 cases of CL are reported in Iran. It is estimated that the actual number of cases is 6-10 times more than that which is reported (2). Golestan Province is one of the important centers of CL in Iran, which is mostly reported from rural populations and the margin of cities (3).
The prevention and control of CL varies depending on the species of the parasite. In endemic areas where the prevalence of CL is high, the correct and timely diagnosis of this disease is of particular importance. Thus, the use of diagnostic methods with high sensitivity and specificity such as PCR is suggested (4). PCR technique is considered a valuable method in epidemiological studies (5). In comparison, current methods for detecting CL such as microscopic examination of Giemsa-stained smears may report as false negative, especially in low parasitic burden (6). Misdiagnosed cases may lead to subsequent consequences for the patient, such as a delay in healing the lesions and the spread of the disease in the region (7).
Therefore, this study aimed to examine Leishmania species in negative ulcer smears from patients suspected of having CL referred to Aq Qala Health Center, Golestan Province, northeastern Iran, using the multiplex nested PCR method.
Methods
This retrospective cross-sectional study was performed on 72 negative ulcer smears from patients suspected of having CL referred to the health center of Aq Qala City from August 2019 to April 2020. These smears had been prepared by Giemsa stain and examined under a light microscope at 400 and 1,000 magnifications. The examiner's final report was negative for detecting Leishmania species.
DNA was extracted from the smears using a Blood DNA isolation kit (DENA Zist Asia, Mashhad, Iran) according to the manufacturer's instructions. The quantity and quality of the extracted DNA were analyzed using a NanoDrop ND-1000 spectrophotometer (PeqLab, Erlangen, Germany).
PrimerPlex v2.6 software was used to design primers for multiplex nested PCR. Two pairs of primers were used for each studied gene. Accordingly, the product of the first reaction was considered as the target of the second reaction. Therefore, two pairs of primers, one for the first reaction and the other for the second reaction, were considered for each of the examined parasites, Leishmania major and Leishmania tropica. Primers were designed for the SSU rDNA gene of the mentioned parasites. Furthermore, DNA for fragments 3 and 4 of histone protein was used to evaluate the accuracy of the multiplex nested PCR reaction as an internal control. The specifications of the primers used are given in Tables 1 and 2.
Multiplex TEMPase 2x Master Mix (Ampliqon, Denmark) was used for this aim according to the manufacturer's instructions. The reactions were performed in a 50 µL final volume, including 25 µL of the 2x Master Mix, 1 µL of each primer, target DNA, and PCR-grade H2O to a total reaction volume of 50 µL. Reaction 1 included 3 steps as follows: 1) denaturation at 94 °C for 2 min, 2) 35 cycles including denaturation at 94 °C for 15 s, annealing at 52.2 °C for 30 s, extension at 72 °C for 30 s, 3) final extension at 72 °C for 2 min. Reaction 2 was carried out in the same way as Reaction 1, except that the annealing was carried out at 50.3 °C.
The PCR product was run on 1.5% agarose gel in TBE buffer. After electrophoresis, the gel was placed in the UV transilluminator, and it was expected to visualize at least two bands related to two internal control genes. If the sample is positive for each of the two parasites under investigation, the bands related to each of those two would be visible. Band lengths are given in Tables 1 and 2.
The number of positive cases of each species of Leishmania major and Leishmania tropica was reported as frequency and percentage.
Results and Discussion
Out of 72 negative smears from the skin ulcers of patients, four (5.55%) samples were positive by multiplex nested PCR. Furthermore, all positive samples were related to Leishmania major species (Figure 1). Out of the four positive cases, three were female, and one was male. The ages of these individuals were 18, 32, 34, and 58 years. All four resided in rural areas and had no prior history of CL. The lesions in each case were located on the ankle, with a single lesion present in each individual. None of the positive cases had any underlying diseases. The duration of the lesions at the time of diagnosis ranged from one to three weeks.
The current method of examining suspected cases of having CL includes the preparation of a smear from the margin of the ulcer and staining with the Giemsa method and microscopic examination. Depending on the expertise of the examiner, the parasite burden of the sample, and the quality of the staining, the results may be associated with false negatives. Therefore, the microscopic examination method has low sensitivity as compared to a highly sensitive molecular method for the diagnosis of CL, such as PCR (8,9).
In a study conducted on 29 microscopically negative samples from patients suspected of having CL, 18 (62%) samples were positive using the PCR method, which shows the superiority of the PCR method compared to the microscopic examination method (10). Moreover, the difference between the number of positive cases in negative samples between the present study (5.55%) and the latter study (62%) shows the significant impact of individual expertise in the microscopic diagnosis of Leishmania. In the present study, all the positive cases were Leishmania major, which was consistent with the results of Hezari et al. in Golestan Province, who reported only Leishmania major species (11). One of the advantages of the PCR method over the microscopic method can be the possibility of determining the parasite species (10).
In another study in Gonbad-e Kavus, out of 65 negative microscopic examination samples from patients suspected of having CL, 34 (53.3%) cases were found to be positive using the PCR method. The positive samples were Leishmania major, similar to the findings of the present study (12).
The results of a study conducted on 62 microscopically negative smears for Leishmania showed that 35 (4.56%) specimens were positive for Leishmania parasite using the PCR method, which is in line with the results of the present study (13). Mohaghegh et al. showed that 1.11% of samples reported as negative microscopically were positive using PCR, which is consistent with the results of the present study; however, the type of parasite found in all positive cases was Leishmania tropica. The difference between the parasite species found in the current study and the latter one is related to the geographical area (14).
In another study, out of 30 negative samples microscopically, 13 (3.43%) samples were positive using the PCR method (15). A comparison among the microscopic method and three different diagnostic methods based on PCR showed that the sensitivity of the microscopic method was 22%, and the sensitivity of the three PCR methods was reported between 64% and 100% (16). In order to decide on the use of different methods for the diagnosis of CL, in addition to the sensitivity and specificity of the methods used, the costs incurred have always been taken into consideration, and the higher cost of the PCR method compared to the microscopic method is one of the considerable points.
The number of positive cases of each species of Leishmania major and Leishmania tropica was reported as frequency and percentage.
Results and Discussion
Out of 72 negative smears from the skin ulcers of patients, four (5.55%) samples were positive by multiplex nested PCR. Furthermore, all positive samples were related to Leishmania major species (Figure 1). Out of the four positive cases, three were female, and one was male. The ages of these individuals were 18, 32, 34, and 58 years. All four resided in rural areas and had no prior history of CL. The lesions in each case were located on the ankle, with a single lesion present in each individual. None of the positive cases had any underlying diseases. The duration of the lesions at the time of diagnosis ranged from one to three weeks.
The current method of examining suspected cases of having CL includes the preparation of a smear from the margin of the ulcer and staining with the Giemsa method and microscopic examination. Depending on the expertise of the examiner, the parasite burden of the sample, and the quality of the staining, the results may be associated with false negatives. Therefore, the microscopic examination method has low sensitivity as compared to a highly sensitive molecular method for the diagnosis of CL, such as PCR (8,9).
In a study conducted on 29 microscopically negative samples from patients suspected of having CL, 18 (62%) samples were positive using the PCR method, which shows the superiority of the PCR method compared to the microscopic examination method (10). Moreover, the difference between the number of positive cases in negative samples between the present study (5.55%) and the latter study (62%) shows the significant impact of individual expertise in the microscopic diagnosis of Leishmania. In the present study, all the positive cases were Leishmania major, which was consistent with the results of Hezari et al. in Golestan Province, who reported only Leishmania major species (11). One of the advantages of the PCR method over the microscopic method can be the possibility of determining the parasite species (10).
In another study in Gonbad-e Kavus, out of 65 negative microscopic examination samples from patients suspected of having CL, 34 (53.3%) cases were found to be positive using the PCR method. The positive samples were Leishmania major, similar to the findings of the present study (12).
The results of a study conducted on 62 microscopically negative smears for Leishmania showed that 35 (4.56%) specimens were positive for Leishmania parasite using the PCR method, which is in line with the results of the present study (13). Mohaghegh et al. showed that 1.11% of samples reported as negative microscopically were positive using PCR, which is consistent with the results of the present study; however, the type of parasite found in all positive cases was Leishmania tropica. The difference between the parasite species found in the current study and the latter one is related to the geographical area (14).
In another study, out of 30 negative samples microscopically, 13 (3.43%) samples were positive using the PCR method (15). A comparison among the microscopic method and three different diagnostic methods based on PCR showed that the sensitivity of the microscopic method was 22%, and the sensitivity of the three PCR methods was reported between 64% and 100% (16). In order to decide on the use of different methods for the diagnosis of CL, in addition to the sensitivity and specificity of the methods used, the costs incurred have always been taken into consideration, and the higher cost of the PCR method compared to the microscopic method is one of the considerable points.
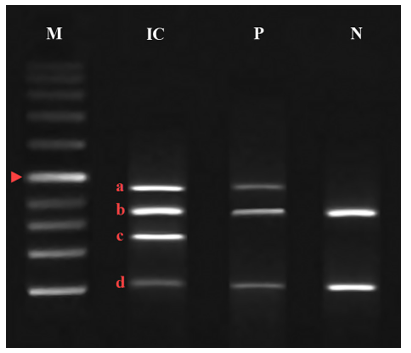 Figure 1. Bands obtained from multiplex nested PCR on negative slides obtained from samples suspected of cutaneous leishmaniasis. M = standard DNA marker (1000 bp); IC = internal control to determine the correct operation of multiplex PCR; P = positive sample; N = negative sample; a = 465 bp band related to Leishmania major; b = 362 bp band related to fragment 3 of histone protein 1; c = 278 bp band related to Leishmania tropica; d = 106 bp band corresponding to fragment 4 of histone protein 1. Arrowhead = 500 bp band |
Conclusion
The results of this study showed the superiority of the multiplex nested PCR method over the microscopic method. In cases suspected of having CL, multiplex nested PCR can be used to avoid inappropriate treatment for patients where the samples are reported negative microscopically. Using the multiplex nested PCR method to determine the type of parasite that causes CL can provide researchers with more accurate information about the prevalence of the disease in epidemiological studies, which helps in making preventive decisions.
Acknowledgement
None.
Funding sources
This research was financially supported by the Vice Chancellor of Research at Golestan University of Medical Sciences. (Grant No: 112237).
Ethical statement
The Ethics Committee of Golestan University of Medical Sciences (Ethical code: IR.GOUMS.REC.1400.166) approved this study. This article has not been published in other journals, nor will it be, and is not under review in any journal.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Author contributions
Fatemeh Livani: Study design; Faramarz Koohsar and Mehdi Khoshrou: Sample collection; Farideh Tohidi: Original draft preparation; Mitra Sharbatkhori, Roghiyeh Faridnia, Ayeneh Hajieh Pangh: Laboratory work; Hamed Kalani: Supervision, Study design, Reviewing and editing; Ganesh Yadagiri: Critical revision.
Data availability statement
All data analyzed in this study are presented in this published article.
Research Article: Brief Report |
Subject:
Parasitology
Received: 2023/09/3 | Accepted: 2023/11/26 | Published: 2025/06/25 | ePublished: 2025/06/25
Received: 2023/09/3 | Accepted: 2023/11/26 | Published: 2025/06/25 | ePublished: 2025/06/25
References
1. Torres-Guerrero E, Quintanilla-Cedillo MR, Ruiz-Esmenjaud J, Arenas R. Leishmaniasis: a review. F1000Res. 2017; 6: 750. [View at Publisher] [DOI] [PMID]
2. Akhtari J, Faridnia R, Kalani H, Bastani R, Fakhar M, Rezvan H, et al. Potent in vitro antileishmanial activity of a nanoformulation of cisplatin with carbon nanotubes against Leishmania major. J Glob Antimicrob Resist. 2019; 2(4):16:11-6. [View at Publisher] [DOI] [PMID] [Google Scholar]
3. Weigle KA, Labrada LA, Lozano C, Santrich C, Barker DC. PCR-based diagnosis of acute and chronic cutaneous leishmaniasis caused by Leishmania (Viannia). J Clin Microbiol. 2002; 40(2): 601-606. [View at Publisher] [DOI] [PMID] [Google Scholar]
4. Aviles H, Belli A, Armijos R, Monroy FP, Harris E. PCR detection and identification of Leishmania parasites in clinical specimens in Ecuador: a comparison with classical diagnostic methods. J Parasitol 1999; 85(2):181-187. [View at Publisher] [DOI] [PMID] [Google Scholar]
5. Mahboudi F, Abolhassani M, Tehrani SR, Azimi M, Asmar M. Differentiation of old and new world leishmania species at complex and species levels by PCR. Scand J Infect Dis. 2002; 34(10): 756-758. [View at Publisher] [DOI] [PMID] [Google Scholar]
6. Pourmohammadi B, Motazedian MH , Hatam GR, Kalantari M, Habibi P, Sarkari B. Comparison of Three Methods for Diagnosis of Cutaneous Leishmaniasis. Iran J Parasitol. 2010; 5(4): 1-8. [View at Publisher] [PMID] [Google Scholar]
7. Thakur Sh, Joshi J, KaurS. Leishmaniasis diagnosis: an update on the use of parasitological, immunological and molecular methods. J Parasit Dis. 2020; 44(2): 253-272. [View at Publisher] [DOI] [PMID] [Google Scholar]
8. Rodrigues E, De Brito M, Mendonca M. Werkhauser RP, Coutinho EM, Souza WV, et al. Evaluation of PCR for diagnosis of Ameri-can cutaneous leishmaniasis in an area of endemicity in Northeastern Brazil. J Clin Microbiol. 2002; 40: 3572-3576. [View at Publisher] [DOI] [PMID] [Google Scholar]
9. Bensoussan E, Nasereddin A, Jonas F, Schnur LF, Jaffe CL. Comparison of PCR assays for diagnosis of cutaneous leishmaniasis. J Clin Microbiol. 2006; 44: 1435-1439. [View at Publisher] [DOI] [PMID] [Google Scholar]
10. Aghamolaei S, Behniafar H, Behravan M, Hajjaran H, Vaziri VM. Probability of false-negative results in microscopical detection of cutaneous leishmaniasis: more accurate screening by kDNA-PCR during epidemiological survey. J Parasit Dis. 2020; 44(4): 781-784. [View at Publisher] [DOI] [PMID] [Google Scholar]
11. Hezari F, Niyyati M, Tabaei SJ, Mohebali M, Vaziri VM, Behniafar H, Azargashb E, Taghipour N. Frequency of cutaneous leishmaniasis and species identification in suspected individuals from Golestan province, Northern Iran in 2014. Iran J Public Health. 2016; 45(10):1348. [View at Publisher] [PMID] [Google Scholar]
12. Pakheh, Abdul Sattar, Fakhar, Sharif, Danesh, Vahid, Ahmadi, Ziba. Epidemiological study of cutaneous leishmaniasis with Leishmania tropica in a new center of Khorasan Razavi province. J Maz Univ Med Sci. 2014; 23 (103): 46-52. [View at Publisher] [Google Scholar]
13. Fakhar M, Mikaeili F, Hatam GR, Habibi P, Karamian M, Motazedian M, Banimostafavi E. A molecular epidemiology survey of cutaneous leishmaniasis in patient referring to parasitology lab at shiraz school of medicine and the importance of PCR assay. Pars J Med Sci. 2014; 8(2):311. [View at Publisher] [DOI] [Google Scholar]
14. Mohaghegh MA, Fata A, Salehi GH, Berenji F. Molecular identification of Leishmania species using samples obtained from negative stained smears. Iran J Parasitol. 2013; 8(2):337. [View at Publisher] [PMID] [Google Scholar]
15. Karamian M, Faroghi Bojd MS, Hemmati M, Saadatjoo A, Barati DA. Molecular identification of cutaneous leishmaniasis agents in Birjand, Iran. J Birjand Univ Med Sci. 2013; 20(2):183-190. [View at Publisher] [Google Scholar]
16. Merdekios B, Pareyn M, Tadesse D, Getu S, Admassu B, Girma N, Leirs H, van Griensven J. Detection of Cutaneous Leishmaniasis Foci in South Ethiopia. Am J Trop Med Hyg. 2021; 51 (2): 79-86. [View at Publisher] [DOI] [PMID] [Google Scholar]
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.